I found mdp on sourceforge. I needed to do PCA for some of my simulations. The mdp.pca function is easy to handle. The input is a m x n matrix, n is the dimension of the space, and m are the observations (rows = observations, cols = dimensions. The doc string is confused as to this point)
The output is a matrix also m x n, but the vectors have been transformed, so that the first column is along the direction with greatest variance etc.
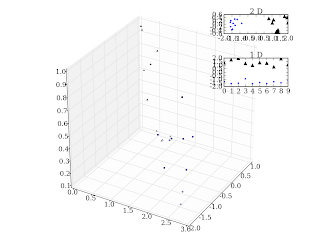
An example is below:
import pylab as m
import matplotlib.axes3d as m3
import mdp
x1 = m.rand(10,3)
x2 = m.rand(10,3) + m.array([[2.,-2.,0.]])
x = m.concatenate((x1,x2))
y = mdp.pca(x)
fig = m.figure()
ax = m3.Axes3D(fig)
ax.scatter3D(x[:10,0].squeeze().T, x[:10,1].squeeze().T, x[:10,2].squeeze().T, marker = '^')
ax.scatter3D(x[10:,0].squeeze().T, x[10:,1].squeeze().T, x[10:,2].squeeze().T, marker = 'o')
m.axis('scaled')
m.axes([.7, .8, .2, .2])
m.plot(y[:10,0], y[:10,1],'k^')
m.plot(y[10:,0], y[10:,1],'b.')
m.axis('scaled')
m.title('2 D')
m.axes([.7, .6, .2, .2])
m.plot(y[:10,0],'k^')
m.plot(y[10:,0],'b.')
m.axis('scaled')
m.title('1 D')
For anyone reading this, the code needs to be updated for the latest version of matplotlib:
ReplyDeleteline 2 should be:
import mpl_toolkits.mplot3d as m3
also add m.show() at the end to display the results.